sklearn_extra.cluster.KMedoids¶
- class sklearn_extra.cluster.KMedoids(n_clusters=8, metric='euclidean', method='alternate', init='heuristic', max_iter=300, random_state=None)[source]¶
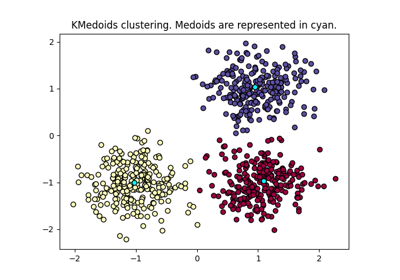
k-medoids clustering.
Read more in the User Guide.
- Parameters:
- n_clustersint, optional, default: 8
The number of clusters to form as well as the number of medoids to generate.
- metricstring, or callable, optional, default: ‘euclidean’
What distance metric to use. See :func:metrics.pairwise_distances metric can be ‘precomputed’, the user must then feed the fit method with a precomputed kernel matrix and not the design matrix X.
- method{‘alternate’, ‘pam’}, default: ‘alternate’
Which algorithm to use. ‘alternate’ is faster while ‘pam’ is more accurate.
- init{‘random’, ‘heuristic’, ‘k-medoids++’, ‘build’}, or array-like of shape
(n_clusters, n_features), optional, default: ‘heuristic’ Specify medoid initialization method. ‘random’ selects n_clusters elements from the dataset. ‘heuristic’ picks the n_clusters points with the smallest sum distance to every other point. ‘k-medoids++’ follows an approach based on k-means++_, and in general, gives initial medoids which are more separated than those generated by the other methods. ‘build’ is a greedy initialization of the medoids used in the original PAM algorithm. Often ‘build’ is more efficient but slower than other initializations on big datasets and it is also very non-robust, if there are outliers in the dataset, use another initialization. If an array is passed, it should be of shape (n_clusters, n_features) and gives the initial centers.
- max_iterint, optional, default300
Specify the maximum number of iterations when fitting. It can be zero in which case only the initialization is computed which may be suitable for large datasets when the initialization is sufficiently efficient (i.e. for ‘build’ init).
- random_stateint, RandomState instance or None, optional
Specify random state for the random number generator. Used to initialise medoids when init=’random’.
See also
KMeansThe KMeans algorithm minimizes the within-cluster sum-of-squares criterion. It scales well to large number of samples.
Notes
Since all pairwise distances are calculated and stored in memory for the duration of fit, the space complexity is O(n_samples ** 2).
References
- Maranzana, F.E., 1963. On the location of supply points to minimize
transportation costs. IBM Systems Journal, 2(2), pp.129-135.
- Park, H.S.and Jun, C.H., 2009. A simple and fast algorithm for K-medoids
clustering. Expert systems with applications, 36(2), pp.3336-3341.
Examples
>>> from sklearn_extra.cluster import KMedoids >>> import numpy as np
>>> X = np.asarray([[1, 2], [1, 4], [1, 0], ... [4, 2], [4, 4], [4, 0]]) >>> kmedoids = KMedoids(n_clusters=2, random_state=0).fit(X) >>> kmedoids.labels_ array([0, 0, 0, 1, 1, 1]) >>> kmedoids.predict([[0,0], [4,4]]) array([0, 1]) >>> kmedoids.cluster_centers_ array([[1., 2.], [4., 2.]]) >>> kmedoids.inertia_ 8.0
See scikit-learn-extra/examples/plot_kmedoids_digits.py for examples of KMedoids with various distance metrics.
- Attributes:
- cluster_centers_array, shape = (n_clusters, n_features)
or None if metric == ‘precomputed’
Cluster centers, i.e. medoids (elements from the original dataset)
- medoid_indices_array, shape = (n_clusters,)
The indices of the medoid rows in X
- labels_array, shape = (n_samples,)
Labels of each point
- inertia_float
Sum of distances of samples to their closest cluster center.
Examples using sklearn_extra.cluster.KMedoids¶
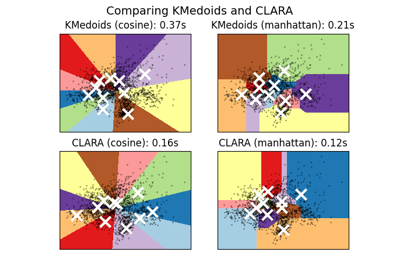
A demo of K-Medoids vs CLARA clustering on the handwritten digits data
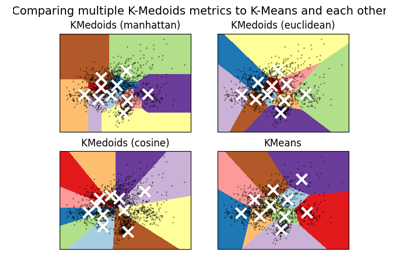
A demo of K-Medoids clustering on the handwritten digits data

A demo of several clustering algorithms on a corrupted dataset