Note
Go to the end to download the full example code
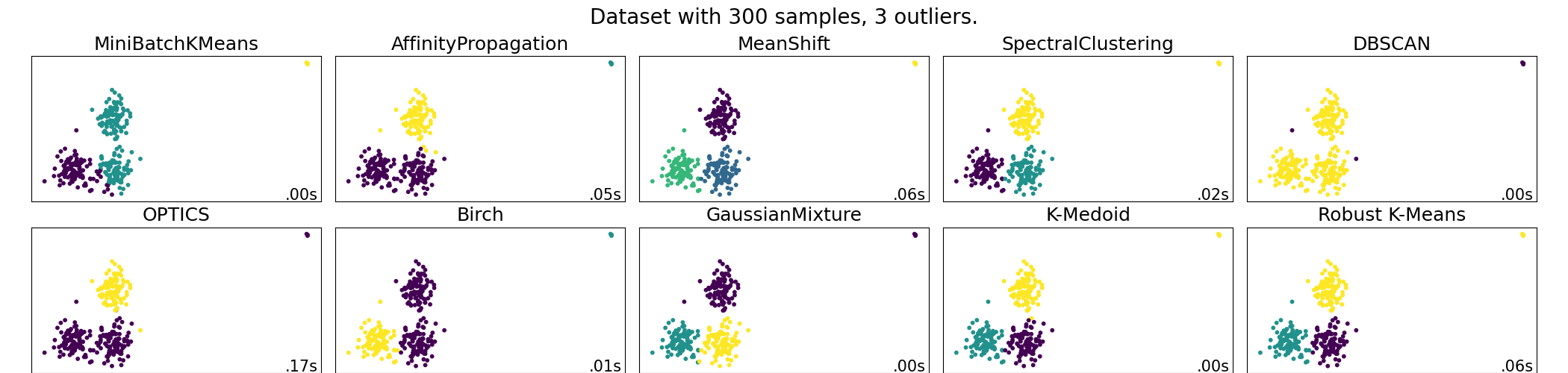
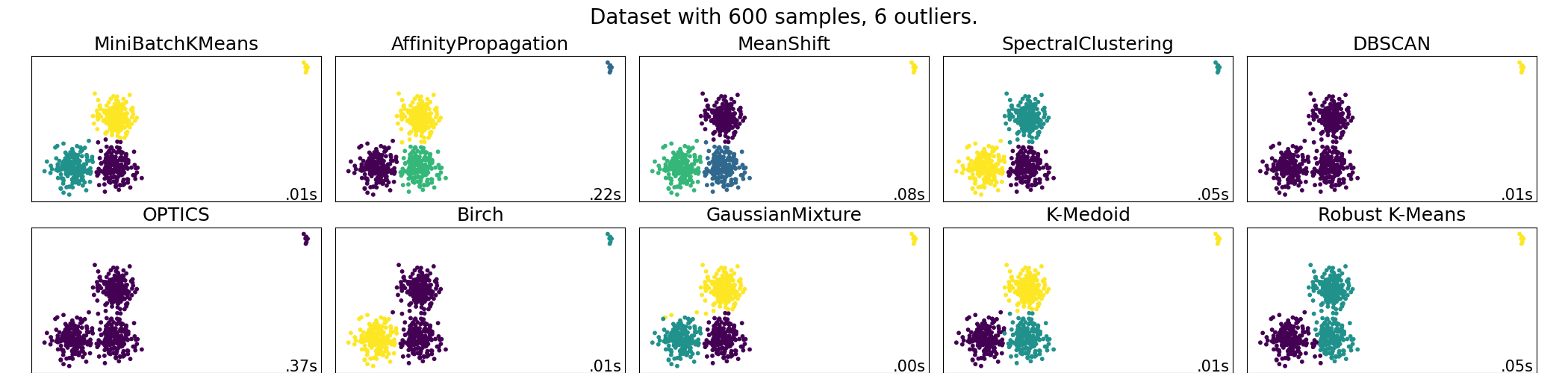
A demo of several clustering algorithms on a corrupted dataset¶
In this example we exhibit the results of various scikit-learn and scikit-learn-extra clustering algorithms on a dataset with outliers. KMedoids is the most stable and efficient algorithm for this application (change the seed to see different behavior for SpectralClustering and the robust kmeans). The mean-shift algorithm, once correctly parameterized, detects the outliers as a class of their own.
/home/docs/checkouts/readthedocs.org/user_builds/scikit-learn-extra/envs/latest/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py:1934: FutureWarning: The default value of `n_init` will change from 3 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
super()._check_params_vs_input(X, default_n_init=3)
/home/docs/checkouts/readthedocs.org/user_builds/scikit-learn-extra/envs/latest/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py:1934: FutureWarning: The default value of `n_init` will change from 3 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
super()._check_params_vs_input(X, default_n_init=3)
/home/docs/checkouts/readthedocs.org/user_builds/scikit-learn-extra/envs/latest/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py:1934: FutureWarning: The default value of `n_init` will change from 3 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
super()._check_params_vs_input(X, default_n_init=3)
/home/docs/checkouts/readthedocs.org/user_builds/scikit-learn-extra/envs/latest/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py:1934: FutureWarning: The default value of `n_init` will change from 3 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
super()._check_params_vs_input(X, default_n_init=3)
print(__doc__)
import time
import numpy as np
import matplotlib.pyplot as plt
from sklearn import cluster, mixture
from sklearn.cluster import KMeans
from sklearn.datasets import make_blobs
from sklearn.utils import shuffle
from sklearn_extra.robust import RobustWeightedKMeans
from sklearn_extra.cluster import KMedoids
rng = np.random.RandomState(42)
centers = [[1, 1], [-1, -1], [1, -1]]
n_clusters = len(centers)
kmeans = KMeans(n_clusters=n_clusters, random_state=rng)
kmedoid = KMedoids(n_clusters=n_clusters, random_state=rng)
two_means = cluster.MiniBatchKMeans(n_clusters=n_clusters, random_state=rng)
spectral = cluster.SpectralClustering(
n_clusters=n_clusters,
eigen_solver="arpack",
affinity="nearest_neighbors",
random_state=rng,
)
dbscan = cluster.DBSCAN()
optics = cluster.OPTICS(min_samples=20, xi=0.1, min_cluster_size=0.2)
affinity_propagation = cluster.AffinityPropagation(
damping=0.75, preference=-220, random_state=rng
)
birch = cluster.Birch(n_clusters=n_clusters)
gmm = mixture.GaussianMixture(
n_components=n_clusters, covariance_type="full", random_state=rng
)
for n_samples in [300, 600]:
# Construct the dataset
X, labels_true = make_blobs(
n_samples=n_samples, centers=centers, cluster_std=0.4, random_state=rng
)
# Change the first 1% entries to outliers
for f in range(int(n_samples / 100)):
X[f] = [10, 3] + rng.normal(size=2) * 0.1
# Shuffle the data so that we don't know where the outlier is.
X = shuffle(X, random_state=rng)
# Define two other clustering algorithms
kmeans_rob = RobustWeightedKMeans(
n_clusters,
weighting="mom",
max_iter=100,
k=int(n_samples / 20),
random_state=rng,
)
bandwidth = cluster.estimate_bandwidth(X, quantile=0.2)
ms = cluster.MeanShift(bandwidth=bandwidth, bin_seeding=True)
clustering_algorithms = (
("MiniBatchKMeans", two_means),
("AffinityPropagation", affinity_propagation),
("MeanShift", ms),
("SpectralClustering", spectral),
("DBSCAN", dbscan),
("OPTICS", optics),
("Birch", birch),
("GaussianMixture", gmm),
("K-Medoid", kmedoid),
("Robust K-Means", kmeans_rob),
)
plot_num = 1
fig = plt.figure(figsize=(9 * 2 + 3, 5))
plt.subplots_adjust(
left=0.02, right=0.98, bottom=0.001, top=0.85, wspace=0.05, hspace=0.18
)
for name, algorithm in clustering_algorithms:
t0 = time.time()
algorithm.fit(X)
t1 = time.time()
if hasattr(algorithm, "labels_"):
y_pred = algorithm.labels_.astype(int)
else:
y_pred = algorithm.predict(X)
plt.subplot(2, int(len(clustering_algorithms) / 2), plot_num)
plt.title(name, size=18)
plt.scatter(X[:, 0], X[:, 1], s=10, c=y_pred)
plt.xticks(())
plt.yticks(())
plt.text(
0.99,
0.01,
("%.2fs" % (t1 - t0)).lstrip("0"),
transform=plt.gca().transAxes,
size=15,
horizontalalignment="right",
)
plt.suptitle(
f"Dataset with {n_samples} samples, {n_samples // 100} outliers.",
size=20,
)
plot_num += 1
Total running time of the script: (0 minutes 1.874 seconds)