Note
Go to the end to download the full example code
A demo of Robust Classification on real dataset “diabetes” from OpenML¶
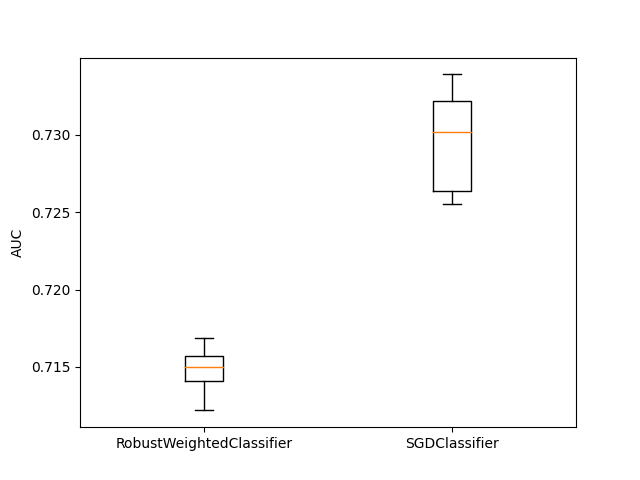
In this example we compare the RobustWeightedCLassifier for classification on the real dataset “diabetes”. We only compare the estimator with SGDClassifier as there is no robust classification estimator in scikit-learn.
/home/docs/checkouts/readthedocs.org/user_builds/scikit-learn-extra/envs/stable/lib/python3.7/site-packages/sklearn/datasets/_openml.py:421: UserWarning: Multiple active versions of the dataset matching the name diabetes exist. Versions may be fundamentally different, returning version 1.
" {version}.".format(name=name, version=res[0]["version"])
Progress: 1 / 10
Progress: 2 / 10
Progress: 3 / 10
Progress: 4 / 10
Progress: 5 / 10
Progress: 6 / 10
Progress: 7 / 10
Progress: 8 / 10
Progress: 9 / 10
Progress: 10 / 10
import matplotlib.pyplot as plt
import numpy as np
from sklearn_extra.robust import RobustWeightedClassifier
from sklearn.linear_model import SGDClassifier
from sklearn.datasets import fetch_openml
from sklearn.metrics import roc_auc_score, make_scorer
from sklearn.model_selection import cross_val_score
from sklearn.preprocessing import RobustScaler
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
X, y = fetch_openml(name="diabetes", as_frame=False, return_X_y=True)
# replace the label names with 0 or 1
y = (y == "tested_positive").astype(int)
# Scale the dataset with sklearn RobustScaler (important for this algorithm)
X = RobustScaler().fit_transform(X)
# Using GridSearchCV, to tune the parameters alpha, eta0, learning_rate, loss
# and average of SGDClassifier, we get the following parameters.
clf_not_rob = SGDClassifier(average=10, learning_rate="optimal", loss="hinge")
# Then, we use this estimator as base_estimator of RobustWeightedEstimator.
# Using GridSearchCV, we tuned the parameters c and eta0, with the
# choice of "huber" weighting because the sample_size is not very large.
clf_rob = RobustWeightedClassifier(
weighting="huber",
loss="hinge",
c=1.35,
max_iter=300,
)
# We compute M times the cross validations in order to also have an estimate
# of the variance of the loss of the estimators.
M = 10
res = []
for f in range(M):
rng = np.random.RandomState(f)
print("\r Progress: %s / %s" % (f + 1, M), end="")
clf = SGDClassifier(
average=10, learning_rate="optimal", loss="hinge", random_state=rng
)
clf_rob = RobustWeightedClassifier(
weighting="huber",
loss="hinge",
c=1.35,
max_iter=300,
random_state=rng,
)
cv_not_rob = cross_val_score(
clf_not_rob, X, y, cv=10, scoring=make_scorer(roc_auc_score)
)
cv_rob = cross_val_score(
clf_rob, X, y, cv=10, scoring=make_scorer(roc_auc_score)
)
res += [[np.mean(cv_rob), np.mean(cv_not_rob)]]
plt.boxplot(
np.array(res), labels=["RobustWeightedClassifier", "SGDClassifier"]
)
plt.ylabel("AUC")
# Remark : when using accuracy score, the optimal hyperparameters change and
# for example the parameter c changes from 1.35 to 10.
plt.show()
Total running time of the script: ( 0 minutes 9.749 seconds)